Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
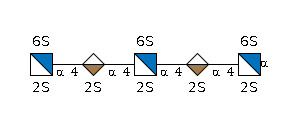
MIRAGE G40789QE
G40789QE
Summary
- GlyTouCan ID
-
G40789QE
- IUPAC Condensed
- GlcN2,6S2(a1-4)IdoA2S(a1-4)GlcN2,6S2(a1-4)IdoA2S(a1-4)GlcN2,6S2(a1-
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 1492.94
3D Structures
GlycoShape
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dglc-HEX-1:5 2s:n-sulfate 3b:a-lido-HEX-1:5|6:a 4s:sulfate 5b:a-dglc-HEX-1:5 6s:n-sulfate 7b:a-lido-HEX-1:5|6:a 8s:sulfate 9b:a-dglc-HEX-1:5 10s:n-sulfate 11s:sulfate 12s:sulfate 13s:sulfate LIN 1:1d(2+1)2n 2:1o(4+1)3d 3:3o(2+1)4n 4:3o(4+1)5d 5:5d(2+1)6n 6:5o(4+1)7d 7:7o(2+1)8n 8:7o(4+1)9d 9:9d(2+1)10n 10:9o(6+1)11n 11:5o(6+1)12n 12:1o(6+1)13n
- WURCS
- WURCS=2.0/2,5,4/[a2122h-1a_1-5_2*NSO/3=O/3=O_6*OSO/3=O/3=O][a2121A-1a_1-5_2*OSO/3=O/3=O]/1-2-1-2-1/a4-b1_b4-c1_c4-d1_d4-e1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 30602608 | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid | Dhindwal S | 2019 Mar 15 |
|
| 15548541 | Crystal Structure of Thrombin Bound to Heparin | Carter WJ | 2005 Jan 28 |
|
| 9655399 | Structure of a heparin-linked biologically active dimer of fibroblast growth factor | DiGabriele AD | 1998 Apr 22 |
|