Table Filtering
Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / Organisms GlycoBase
GlycoBase
GlycoBase is a relational database which contains the HPLC elution positions for over 350 2-AB labelled N-glycan structures together with predicted products of exoglycosidase digestions.
GlycoBase and autoGU: tools for HPLC-based glycan analysis
Campbell MP, et al.
Bioinformatics. 2008 May 1;24(9):1214-6. PMID: 18344517
| Source | Last Updated |
|---|---|
| Glycobase | April 18, 2024 |
| GlyTouCan ID | GlycoBase ID ▼ | Compositions | Compositions (GlyTouCan ID) | Mass | GU (average) | GU (standard deviation) |
|---|---|---|---|---|---|---|
 G73216PG
G73216PG
|
27850 | Hexose3NeuGc2 | 1118.314453125 | 6.340000152587891 | 38.04001235961914 | |
| 27849 | HexNAc1Hexose3NeuGc1 |
 G72126TO
G72126TO
|
1014.3035278320312 | 5.364999771118164 | 16.095020294189453 | |
| 27848 | Hexose2NeuGc2 |
 G07529OK
G07529OK
|
956.2667846679688 | 5.260000228881836 | 42.080623626708984 | |
 G34163VP
G34163VP
|
27847 | Hexose3NeuGc1 | 811.2288208007812 | 4.15500020980835 | 4.1550750732421875 | |
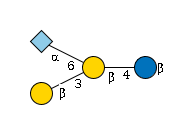 G38048SB
G38048SB
|
27846 | Hexose3NeuGc1 | 811.2288208007812 | 4.394999980926514 | 13.185009002685547 | |
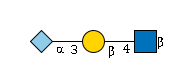 G08169IN
G08169IN
|
27845 | HexNAc1Hexose1NeuGc1 |
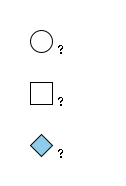 G47967RK
G47967RK
|
690.2081298828125 | 3.3685715198516846 | 20.211429595947266 |
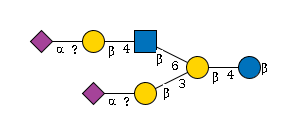 G79342GW
G79342GW
|
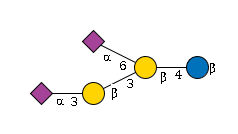
27843 | HexNAc1Hexose4NeuAc2 |
 G21193WJ
G21193WJ
|
1451.4449462890625 | 7.183750152587891 | 50.286800384521484 |
| 27842 | HexNAc2Hexose4NeuAc1 |
 G91331SN
G91331SN
|
1363.429931640625 | 6.827777862548828 | 54.622230529785156 | |
| 27841 | HexNAc2Hexose4NeuAc1 |
 G91331SN
G91331SN
|
1363.429931640625 | 6.434999942779541 | 45.045013427734375 | |
 G05712GH
G05712GH
|
27838 | Hexose3NeuAc2 |
 G03084ZT
G03084ZT
|
1086.322509765625 | 5.476666450500488 | 43.8133430480957 |
| 27837 | Hexose2NeuAc1NeuGc1 |
 G75441FF
G75441FF
|
940.2708129882812 | 4.712500095367432 | 32.987510681152344 | |
 G39838GA
G39838GA
|
27835 | Hexose3NeuAc1 |
 G34081LT
G34081LT
|
795.2327880859375 | 4.002307891845703 | 48.02769470214844 |
 G82385GE
G82385GE
|
27834 | Hexose2NeuGc1 |
 G27785LA
G27785LA
|
649.1810913085938 | 3.9100000858306885 | 31.28000831604004 |
 G24684AO
G24684AO
|
27833 | HexNAc1Hexose1NeuGc1 |
 G47967RK
G47967RK
|
690.2081298828125 | 3.7216665744781494 | 18.60833740234375 |
| 27832 | HexNAc1Hexose1NeuAc1 |
 G74722FL
G74722FL
|
674.2120971679688 | 3.0446152687072754 | 36.53538513183594 | |
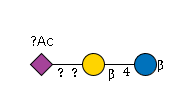 G51304WT
G51304WT
|
27831 | Hexose2NeuAc1 |
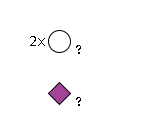 G17756LB
G17756LB
|
633.1851196289062 | 2.454444408416748 | 19.635587692260742 |
| 27830 | HexNAc6Hexose7NeuAc4 |
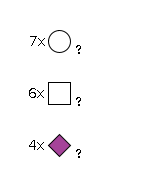 G66088HZ
G66088HZ
|
3535.140869140625 | 13.092727661132812 | 130.9285125732422 | |
| 27829 | Fuc1HexNAc6Hexose7NeuAc3 |
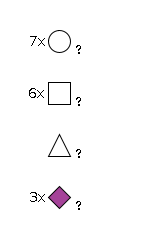 G39595FH
G39595FH
|
3390.10400390625 | 12.616000175476074 | 113.54399871826172 | |
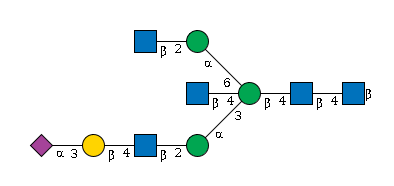
| 27828 | Fuc2HexNAc6Hexose7NeuAc1 |
 G74728JK
G74728JK
|
2953.977294921875 | 12.267999649047852 | 110.41200256347656 | |
| 27827 | HexNAc7Hexose7NeuAc3 |
 G99608DG
G99608DG
|
3447.1259765625 | 12.267999649047852 | 110.41200256347656 | |
| 27826 | Fuc3HexNAc6Hexose7 |
 G22276PO
G22276PO
|
2808.940185546875 | 12.431500434875488 | 236.19850158691406 | |
 G55015QS
G55015QS
|
27825 | HexNAc5Hexose4NeuAc1 |
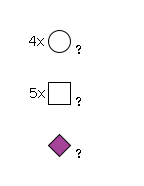 G24835MQ
G24835MQ
|
1972.654052734375 | 7.923999786376953 | 71.31600189208984 |
| 27823 | Fuc2HexNAc5Hexose6NeuAc3 |
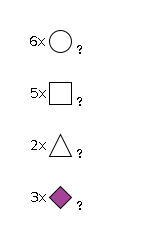 G55412XP
G55412XP
|
3171.0341796875 | 12.479000091552734 | 112.31099700927734 | |
| 27822 | Fuc1HexNAc5Hexose6NeuAc3 |
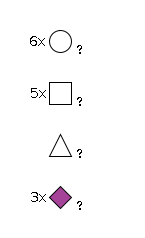 G15169WU
G15169WU
|
3024.9814453125 | 12.027999877929688 | 108.25199890136719 | |
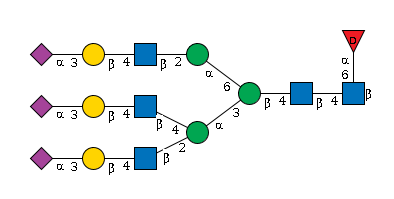 G99204QC
G99204QC
|
27820 | Fuc1HexNAc5Hexose6NeuAc3 |
 G15169WU
G15169WU
|
3024.9814453125 | 11.26911735534668 | 371.8812255859375 |
