Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G00035MO
G00035MO
Summary
- GlyTouCan ID
-
G00035MO
- IUPAC Condensed
- GlcNAc(b1-3)GalNAc(a1-
- Motifs
- O-Glycan core 3
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 424.17
3D Structures
- GlycoNAVI
- G00035MO
- GlycoShape
- G00035MO
- GLYCAM
- DGlcpNAcb1-3DGalpNAca1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Homo sapiens (human) | |
| Rattus norvegicus (Norway rat) |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| ST6GALNAC1 | CMP-Neu5Ac |
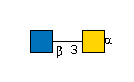 G00035MO
G00035MO
|
O-glycan Synthesis |
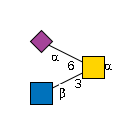 G63334FZ
G63334FZ
|
O-glycan Synthesis | |
| B4GALT4 | UDP-Gal |
 G00035MO
G00035MO
|
p-Nitrophenyl |
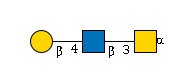 G32723SL
G32723SL
|
p-Nitrophenyl | |
| B3GALT5 | UDP-Gal |
 G00035MO
G00035MO
|
a-OpNp |
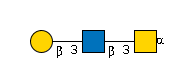 G11647UD
G11647UD
|
a-OpNp | |
| B4GALT1 | UDP-Gal |
 G00035MO
G00035MO
|
O-glycan Synthesis |
 G32723SL
G32723SL
|
O-glycan Synthesis | |
| B3GALT5 | UDP-Gal |
 G00035MO
G00035MO
|
O-glycan Synthesis |
 G11647UD
G11647UD
|
O-glycan Synthesis |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| C1GALT1 | UDP-Gal |
 G00035MO
G00035MO
|
O-glycan Synthesis | |
| B3GNT2 | UDP-GlcNAc |
 G00035MO
G00035MO
|
[alpha]1-OBn | |
| A4GNT | UDP-GlcNAc |
 G00035MO
G00035MO
|
pNP | |
| GCNT1 | UDP-GlcNAc |
 G00035MO
G00035MO
|
[alpha]-1-p-nitrophenyl | |
| B3GNT6 | UDP-GlcNAc |
 G00035MO
G00035MO
|
[alpha]-pNP(core 3) |
KEGG BRITE Database
Core Protein
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl 3b:b-dglc-HEX-1:5 4s:n-acetyl LIN 1:1d(2+1)2n 2:1o(3+1)3d 3:3d(2+1)4n
- WURCS
- WURCS=2.0/2,2,1/[a2112h-1a_1-5_2*NCC/3=O][a2122h-1b_1-5_2*NCC/3=O]/1-2/a3-b1
PubAnnotation
 G00037MO
G00037MO
 G57321FI
G57321FI
