Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G57321FI
G57321FI
Summary
- GlyTouCan ID
-
G57321FI
- IUPAC Condensed
- GalNAc(a1-
- Motifs
- Tn antigen
External Links
3D Structures
- GlycoNAVI
- G57321FI
- GlycoShape
- G57321FI
- GLYCAM
- DGalpNAca1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Severe acute respiratory syndrome coronavirus 2 | |
| Saccharomyces cerevisiae (brewer's yeast) | |
| Bos taurus (domestic cattle) | |
| Rattus norvegicus (Norway rat) | |
| Mesocestoides corti |
Taxonomic Hierarchy
root
cellular organisms [inference]
Eukaryota (eucaryotes) [inference]
Opisthokonta [inference]
Metazoa (metazoans) [inference]
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | (not applicable) |
 G57321FI
G57321FI
|
Ser/Thr |
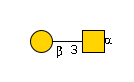 G00031MO
G00031MO
|
Ser/Thr | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
O-glycan Synthesis |
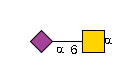 G36123IU
G36123IU
|
O-glycan Synthesis | |
| ST6GALNAC1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
 G36123IU
G36123IU
|
Ser/Thr | |
| B3GNT6 | UDP-GlcNAc |
 G57321FI
G57321FI
|
[alpha]-pNP |
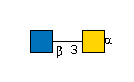 G00035MO
G00035MO
|
[alpha]-pNP |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| C1GALT1 | UDP-Gal |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G00031MO
G00031MO
|
O-glycan Synthesis | |
| C1GALT1 | UDP-Gal |
 G57321FI
G57321FI
|
PSGL-1(EYEYLDYDFLPETEPPEM) |
 G00031MO
G00031MO
|
PSGL-1(EYEYLDYDFLPETEPPEM) | |
| B3GNT6 | UDP-GlcNAc |
 G57321FI
G57321FI
|
O-glycan Synthesis |
 G00035MO
G00035MO
|
O-glycan Synthesis | |
| B3GNT6 | UDP-GlcNAc |
 G57321FI
G57321FI
|
[alpha]-pNP |
 G00035MO
G00035MO
|
[alpha]-pNP |
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| ST8SIA6 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
||
| ST6GALNAC2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
para-nitrophenol | |
| ST6GAL2 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
para-nitrophenol | |
| ST6GAL1 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
para-nitrophenol | |
| ST6GALNAC3 | CMP-Neu5Ac |
 G57321FI
G57321FI
|
Ser/Thr |
KEGG BRITE Database
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P08174 | Complement decay-accelerating factor | ||
| P08476 | Inhibin beta A chain | ||
| P08572 | Collagen alpha-2(IV) chain | ||
| P08575 | Receptor-type tyrosine-protein phosphatase C | ||
| P08581 | Hepatocyte growth factor receptor | ||
| P08648 | Integrin alpha-5 | ||
| P08887 | Interleukin-6 receptor subunit alpha | ||
| P08949 | Neuromedin-B | ||
| P09603 | Macrophage colony-stimulating factor 1 | ||
| P09758 | Tumor-associated calcium signal transducer 2 |
GlycoEpitope
- Epitope ID
- EP0021
- Epitope Name
- Tn Antigen
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O101 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O107 | Escherichia coli |
| E.coli O112ab | Escherichia coli |
| E.coli O112ac | Escherichia coli |
| E.coli O116 | Escherichia coli |
| E.coli O117 | Escherichia coli |
| E.coli O125ab | Escherichia coli |
| E.coli O127 | Escherichia coli |
| E.coli O128ab-H27 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dgal-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2112h-1a_1-5_2*NCC/3=O]/1/
Literature (from GlyTouCan)
- BCSDB
-
- GlycoEpitope
-
PubAnnotation
| Title | Authors | Source | Publication Date | PubMed ID |
|---|---|---|---|---|
| GALNTs: master regulators of metastasis-associated epithelial-mesenchymal transition (EMT)? |
|
Glycobiology | 2022 Jun 13 | 35312770 |
| The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors. |
|
J Biol Chem | 2017 Jul 21 | 28546425 |
| The Quest for Anticancer Vaccines: Deciphering the Fine-Epitope Specificity of Cancer-Related Monoclonal Antibodies by Combining Microarray Screening and Saturation Transfer Difference NMR. |
|
J Am Chem Soc | 2015 Oct 7 | 26366611 |
| Single molecule study of heterotypic interactions between mucins possessing the Tn cancer antigen. |
|
Glycobiology | 2015 May | 25527429 |
| Loss of Core 1-derived O-Glycans Decreases Breast Cancer Development in Mice. |
|
J Biol Chem | 2015 Aug 14 | 26124270 |
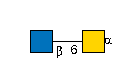 G00041MO
G00041MO
